Gamma mutagenesis to identify diagnostic markers for heat tolerance in wheat
Nick Collins, University of Adelaide
Background
This project focused on the WtmsDW locus of wheat, which controls natural variation in heat stress tolerance – specifically relating to the pollen sterility caused by heat exposure at the booting stage of development (Erena et al. 2021). The aim was to generate overlapping deletion mutations across the WtmsDW region on chromosome 2B using gamma irradiation of seeds, to enable more accurate localisation of WtmsDW. This was to allow closer DNA markers to be identified for use by breeders, and help facilitate the identification (cloning) of the WtmsDW heat tolerance gene.
Results and Discussion
The line used in mutagenesis was ‘NIL-T’ carrying the WtmsDW heat tolerance allele from cv. Waagan (Erena et al. 2021). Seed were given four levels of gamma irradiation from a 60Co source (~196, 229, 262 and 294 Gy; ChemCentre, Perth), then sub-samples sown to assess dose effects. The 262 and 294 Gy dosages were chosen for full population development, based on (a) stunting of M1 seedlings (~31–34% relative to control; Fig. 1) being similar to that observed in the successful (200 Gy) mutagenesis treatment of Mago et al. (2004), (b) similarity of these doses to the 300 Gy used by Draeger et al. (2020) to generate deletion mutants, (c) a recommended dose range of 150-350 Gy for mutagenesis of T. aestivum (FAO/IAEA, 2018), and (d) subsequent healthy appearance of the vast majority of M1 plants.

Figure 1. Seedling height (soil to tip of first green leaf), two weeks after
sowing, in M1 plants derived from irradiated seed. Chart shows means ±
S.E., based on five replicates of eight plants each.
Approximately 300 M1 plants were grown for each of the two chosen dosages (262 and 294 Gy) in pots in a greenhouse. At maturity, one to five spikes were harvested separately per plant (main stems and primary tillers only), to give 1,445 M2 seed packets. Each of these spikes represents an independent sample for mutation, since (based anatomy of the embryo in the seed; Kirby 2002) they would have arisen from separate cell groups present at the time of mutagenesis.
Two to three M2 seed from each M1 spike were sown and leaf
samples taken from these M2 plants (and some control genotypes) for DNA extraction. DNA was extracted from freeze-dried tissue using an SDS-based extraction buffer. At maturity, seed from each fertile plant was harvested, giving 3,093 M3 seed packets.
M2 plants appeared generally healthy and few exhibited clear defects, consistent with known genetic compensation effects between the triplicate genomes of hexaploid wheat (Sears 1966), e.g., only 27 M2 seeds failed to germinate, 13 seedlings were too small or sick to sample, and 38 plants were completely sterile (set no seed).
Genetic markers based on KASPTM assays (Semagn et al. 2014) were used to screen the M2 DNAs for deletions. Initially, five markers roughly evenly spaced through the 4.8 Mb (0.02 cM) WtmsDW genetic interval on chromosome arm 2BS (markers m4, Wtm4, Wtm10, Wtm15 and Wtm16) were used to screen all 3,093 M2 DNAs. For those that looked promising, seed from the corresponding M3 seed packets were sown, and the resulting M3 plants analysed using all 35 markers.
In total, 12 independent deletion mutants for the WtmsDW interval were validated by recovering homozygote mutants in the M3 generation (Fig. 2). Five of these (D39, D185, D297, D111 and C169) were discovered as homozygous deletion mutant M2 plants, while the other seven were detected as potential ‘hemizygote’ M2 plants (only one chromosome copy with a deletion, the other being wild-type) and were then recovered as homozygote deletion mutants in the M3 (explained in more detail in Appendix A).
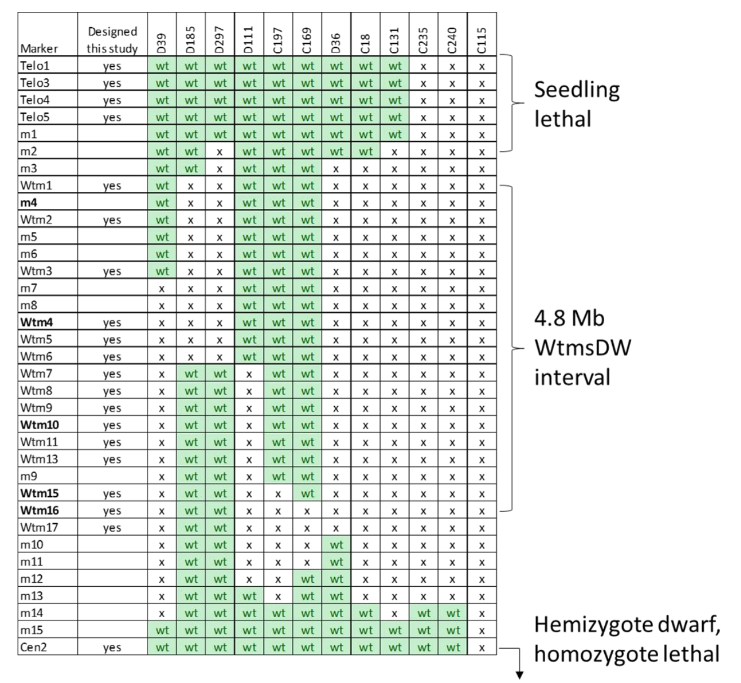
Figure 2. Summary of deletion mutants identified for the WtmsDW locus on the short arm of wheat chromosome 2B. wt, wild-type marker allele; x, deleted marker. The five markers used to screen the whole M2 population are in bold. Telo and Cen markers are close to the 2BS telomere and centromere, respectively. Genomic coordinates of markers on 2BS withheld for reasons of confidentiality.
Six of the 12 mutants had deletion breakpoints within the 4.8 Mb WtmsDW genetic interval, and six had the whole WtmsDW interval deleted (Fig. 2). Three deletions seemed likely to extend all the way to the end (telomere) of the 2BS arm. One of these (C115) essentially encompassed the whole 2BS chromosome arm and may represent a nullisomic 2B line missing the entire chromosome.
Some of the M2 plants with homozygous deletions spanning the whole WtmsDW interval appeared normal and were fully fertile (set seed). However, loci giving dwarf or lethal effects when deleted were defined above and below WtmsDW (Fig. 2; and described in Appendix B).
All of the deletion mutants (isolated in the tolerant background) were still tolerant (Fig. 3), despite the fact that the deletions collectively covered the entire WtmsDW genetic interval (Fig. 2). Our conclusion was that the WtmsDW locus effect derives from an intolerane gene function that is present only in the intolerant line where it is damaging to anthers under heat stress (i.e., a ‘toxic’ gene under heat stress); deletion of the WtmsDW locus in the tolerant NIL-T line did not alter fertility as the toxic gene is not present in this line.

Figure 3. All homozygous deletion mutants were tolerant. Shown are means ±S.E. of grain set % at the two lowest floret positions on the spikelets, on the main stem, after heat stress (37 °C/27 °C day/night, for three days) applied at booting (when bases of the last two leaf blades were 6 cm apart), or under non-stress control conditions. The first nine mutants from Fig. 2 are represented (in the same order), plus the intolerant NIL (NIL-I) and tolerant NIL (NIL-T; the line that was mutated). Only NIL-I was intolerant. For those lines that had control data, N=6 (control) or 12 (heat). For the others (heat data only), N=1–4.
Fortunately, what this meant is that the current mutants are not useful for narrowing down the WtmsDW interval, cloning the gene and identifying closer markers. To achieve these aims, the whole mutant screening process will need to be repeated in the intolerant (NIL-I) background. This is now being undertaken in the newly funded ARC Linkage project LP210301062. Experience, information and some markers arising in the Yitpi project will make this easier the second time around.
The mutant population generated in the NIL-T line represents a resource that could be used to identify deletion mutants for other loci of interest, since the M2 and M3 seed, plus the M2 DNAs, are in storage. This represents another positive outcome for the Yitpi project.
Publication and extension
Findings of this Yitpi project will ultimately be published (with acknowledgment of funding from Yitpi Foundation), when the mutagenesis is repeated in the NIL-I background, and progress is made in cloning WtmsDW and identifying improved markers. Information on improved markers will be emailed to all the Australian wheat breeders as soon as it is available, to allow practical benefits of the work to be realised as soon as possible.
Appendix A. Notes on marker design and screening strategy
Previous studies using KASP assays to screen for deletion mutants in wheat (e.g., Draeger et al. 2020) did not describe how the markers were designed. However, such a description might be useful for others wanting to use this approach. KASP assays typically use two ‘allele-specific’ primers and a common primer oriented in the other direction. Fluorescent dyes (FAM or HEX) become activated when the allele-specific primer(s) find a perfect template and become incorporated into double-stranded PCR products (Fig. 4).
Markers used in the Yitpi project were of two types (Fig. 5). Type 1 were ‘conventional’ single locus co-dominant KASP markers sourced from previous work to map WtmsDW in a Waagan (tolerant) x Drysdale (intolerant) cross.
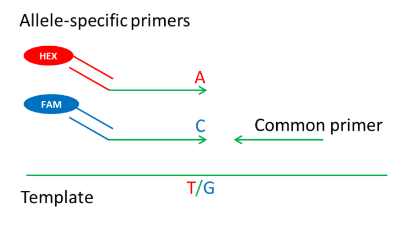
Figure 4. Schematic of a KASP assay
Homozygous deletion mutants in NIL-T (carrying the Waagan WtmsDW allele) gave no PCR signal so resembled negative control (no DNA template) samples on the fluorescence plot (Fig. 5A). Hemizygote plants (in which one 2BS chromosome copy had a deletion and the other copy was wild-type) could not be identified using this marker type (resembled wild-type NIL-T), as the KASP PCR reaction is not quantitative. In Type 2 markers, one ‘allele specific’ primer was specific to a gene in the WtmsDW region on 2B, while the other was designed to match homoeologous gene copies present on 2A and/or 2D. Homozygous deletion mutants in NIL-T appeared as a fluorescence signal only in the 2A/2D dimension (Fig. 5B). The 2A/2D primer seemed to compete with the 2B primer for limiting reaction components (e.g., dNTPs), such that hemizygote WtmsDW mutant plants could be identified by a reduction in the ratio of 2B signal vs. 2A/2D signal (Fig. 5B).

Figure 5. KASP fluorescence plots generated using KrakenTM software (LGC Genomics, UK), representing M3 plants and control genotypes, for Type 1 marker m4 (A) and Type 2 marker Wtm16 (B). Each dot represents a single DNA sample (plant), with position in the X- and Y- axes showing signal intensity from the respective ‘allele specific’ primer fluorophores FAM and HEX. For this figure, dot colours were manually assigned according to the labelled genotypes deduced from all the available information on these plants. See text for further explanation.
In mutant sectors of M1 spikes, M2 seed were expected to segregate in a 1:2:1 ratio of wild-type : hemizygote : homozygous deletion mutant. Therefore, theoretically, use of Type 2 markers to detect hemizygotes could triple the efficiency of mutant detection. In practice however, the proximity of the wild type NIL-T and hemizygote mutant clusters (Fig. 5B) led to some hemizygote miscalls (false positive or false negatives). The accuracy of the hemizygote identification was increased by selecting only those lines showing hemizygote calls for adjacent markers – a strategy that reduced false positives to an estimated 0.3– 2.0% of plants from these clusters, based on results of the M3 validation work (data not shown).
The four Type 2 markers used in the initial screen identified at least 220 other putative hemizygous M2 plants (mostly single marker hemizygote calls) that have not yet been followed up on for validation. However, no more than a handful of these would be expected to represent genuine mutants, given that hemizygote M2s should only be twice as frequent as the homozygotes (of which there were 5; and 7 hemizygotes were already validated).
Appendix B. Dwarf/lethal mutant phenotypes discovered
All three terminal deletions recovered as homozygotes (C235, C240 and C115) showed a recessive extreme dwarf mutant phenotype, defining gene(s) required for healthy wheat seedling growth located in the top quarter (69 Mb) of 2BS, distal of marker m2 (Fig. 2). These seedlings grew no taller than ~4 cm and died within two months. This was unexpected, given that the nullisomic of cv. Chinese Spring missing the whole chromosome 2B was reported to be a viable plant able to grow fertile spikes (Sears 1966). Such mutants may have been among the aforementioned 13 M2 seedlings that were too small or sick to sample for DNA.
There also appeared to be gene(s) on the long arm of chromosome 2B (2BL) that gave dwarfed plants (~40% reduced height at maturity, but otherwise healthy) when in hemizygous deleted state, and no seed recovered in homozygous deleted state. This was deduced because: (a) there were six M3 families segregating hemizygous:wild-type for deletions of the whole of the WtmsDW locus, in which the hemizygotes were dwarfs, but from which no homozygous deletion plants were recovered, despite 100% seed germination, and (b) there was a deletion spanning virtually the whole of 2BS (C115_1; Fig. 2) that did not show these phenotypes. The absence of homozygous deletion seed may reflect a gametophyte lethal effect in pollen and/or the embryo sac. WtmsDW mutants showing these phenotypes may have represented loss of the entire 2B chromosome. Similar to the terminal 2BS deletions, this information seemed inconsistent with Sears’ (1966) report that 2B nullisomic plants could be healthy.
References
Draeger T, et al. (2020). Dmc1 is a candidate for temperature tolerance during wheat meiosis. Theoretical and Applied Genetics 133:809–828.
Erena MF, et al. (2021). The WtmsDW locus on wheat chromosome 2B controls major natural variation for floret sterility responses to heat stress at booting stage. Frontiers in Plant Science 12:635397.
FAO/IAEA (2018). Manual on Mutation Breeding – Third edition. Spencer-Lopes MM, Forster BP and Jankuloski L (eds.), Food and Agriculture Organization of the United Nations. Rome, Italy. 301 pp.
Kirby EJM (2002). Botany of the wheat plant. In Bread Wheat Improvement and Production (Curtis BC, Rajaram S, Gómez Macpherson H, eds.). Food and Agriculture Organization of the United Nations (FAO), Rome, Italy. https://www.fao.org/3/Y4011e/y4011e05.htm
Mago R, et al. (2004). Resistance genes for rye stem rust (SrR) and barley powdery mildew (Mla) are located in syntenic regions on short arm of chromosome. Genome 47:112–121.
Sears ER (1966). Nullisomic-tetrasomic combinations in hexaploid wheat, pp 29-45 in Chromosome Manipulation and Plant Genetics, edited by Riley R and Lewis KR. Oliver & Boyd, Edinburgh.
Semagn K et al. (2014). Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP): overview of the technology and its application in crop improvement. Molecular Breeding 33:1–14.